Single-voxel MRS datasets
This is a list of single-voxel MRS datasets.
2SPECIAL vGRAPPA
This algorithm decomposes simultaneously acquired multi-voxel data to their respective origin regions. Based on the split-slice GRAPPA code by Steen Moeller, University of Minnesota, USA.
| 2SPECIAL vGRAPPA | |
|---|---|
| Developer | Layla Tabea Riemann, M.Sc. (Physikalisch-Technische Bundesanstalt, PTB) |
| Format | SIEMENS DAT |
| Sequence | (2)SPECIAL |
| License | CC BY-NC-SA 4.0 |
| Credit | Please cite the following publication if you use the vGRAPPA algorithm: L.T.Riemann, C.S.Aigner, R.Mekle, S.Schmitter, B.Ittermann, A.Fillmer; Fourier-based decomposition approach for simultaneous acquisition of 1H spectra from two voxels in vivo at short echo times, Proc. Int. Soc. Magn. Reson. Med. 2021:007 |
| Contact | layla.riemann@ptb.de |
Basis sets for PRESS, STEAM and sLASER for multiple vendors
These basis sets were generated using the exact waveforms, timings and 128^3 spatial points to appropriately accommodate the sidebands for each vendor/sequence combination. More details about the acquisition can be found in the reference below.
| Basis sets for PRESS, STEAM and sLASER for multiple vendors | |
|---|---|
| Developer | Karl Landheer, Kelley Swanberg, Christoph Juchem |
| Format | Both .RAW and .mat (INSPECTOR, readable in MATLAB) are provided. All files were generated using MATLAB |
| Sequence | PRESS, sLASER, STEAM |
| License | BSD3 |
| Credit | Please cite the following publication if you use this dataset: Landheer K, Swanberg, K, Juchem C. Magnetic resonance Spectrum simulator (MARSS), a novel software package for fast and computationally efficient basis set simulation. NMR Biomed. 2019, e4219. doi.org/10.1002/nbm.4129 |
| Contact | cwj2112@columbia.edu |
Synthetic brain MR spectroscopy data - sLASER - TE: 35ms - 3T
Synthetic MR spectroscopy data for quantification purposes (included ground-truth concnetration levels) of use for benchmarking (model fitting, machine learning or neural networks algorithms) or of use in deep learning contexts for pre-training, fine-tuning or ablation study. Various dataset size and distribution of concentrations for brain spectra are included (for full description, see README.md file available at the URL reported below or at https://github.com/bellarude/MRS_detasets). Variation of shim quality, SNR levels and MMBG contribution are included. Spectra are free of 0th- and 1st-order phase offset as well as freqeuncy drifts. (optional) Additional downscaled water signal is added (for some datasets) for quantification referencing. (otpional) Spectral preprocessing to spectrogram is included. For further details see: https://doi.org/10.1002/mrm.29561
| Synthetic brain MR spectroscopy data - sLASER - TE: 35ms - 3T | |
|---|---|
| Developer | Rudy Rizzo, MSc and Roland Kreis, PhD |
| Format | Matlab .dat |
| Sequence | sLASER, TE 35ms, 3T |
| License | CC-BY 4.0 (https://creativecommons.org/licenses/by/4.0/legalcode) |
| Credit | Please cite Rizzo R. et al, Quantification of MR spectra by deep learning in an idealized setting: Investigation of forms of input, network architectures, optimization by ensembles of networks, and training bias, Magn. Reson. Med. 89(5):1707-1727 (2023). |
| Contact | roland.kreis@insel.ch |
Big GABA

This repository contains a subset of a large-scale multi-vendor, multi-site collection of single-voxel MRS and structural MRI datasets that were acquired internationally across 26 research sites on 3T MRI scanners from the three major vendors (GE, Philips, Siemens). Each site acquired up to 12 datasets.
| Big GABA | |
|---|---|
| Developer | Richard AE Edden |
| Format | SDAT, TWIX, P |
| Sequence | MEGA-PRESS, PRESS |
| License | open-source |
| Credit | If using these data, please cite: (Mikkelsen M et al. Big GABA: Edited MR spectroscopy at 24 research sites. NeuroImage 2017;159:32–45. doi:10.1016/j.neuroimage.2017.07.021), (Mikkelsen M et al. Big GABA II: Water-referenced edited MR spectroscopy at 25 research sites. NeuroImage 2019;191:537–548. doi:10.1016/j.neuroimage.2019.02.059), and (Povazan M et al. Comparison of multivendor single-voxel MR spectroscopy data acquired in healthy brain at 26 sites. Radiology 2020. doi:10.1148/radiol.2020191037), and acknowledge NIH grant R01 EB016089. |
| Contact | gabamrs@gmail.com |
Fitting challenge data, MRS Workshop 2016

Synthetic data mimicking single-voxel 3 T PRESS TE = 30 ms MRS spectra. Data were simulated using ideal pulses and the known metabolites concentrations in the human brain. The experimentally acquired macromolecular contribution was also used. The following things were modulated in the datasets: non-Lorentzian broadening, concentrations of GABA, GSH and macromolecules, signal-to-noise (SNR) variations, and artifacts. These data was used in the Fitting Challenge organized as part of the ISMRM Workshop on MR Spectroscopy: From Current Best Practice to Latest Frontiers, Lake Constance, Germany, 2016.
| Fitting challenge data, MRS Workshop 2016 | |
|---|---|
| Developer | Malgorzata Marjanska, Dinesh Deelchand, Roland Kreis |
| Format | text, LCModel (.RAW and .h2o), and jMRUI (ascii text) |
| Sequence | PRESS TE = 30 ms, simulated |
| License | BSD3 |
| Credit | Please acknowledge developers as listed above and list following URL: https://www.ismrm.org/workshops/Spectroscopy16/mrs_fitting_challenge/ |
| Contact | gosia@umn.edu |
MM human brain spectra 9.4T - TE series / Fit Settings LCModel
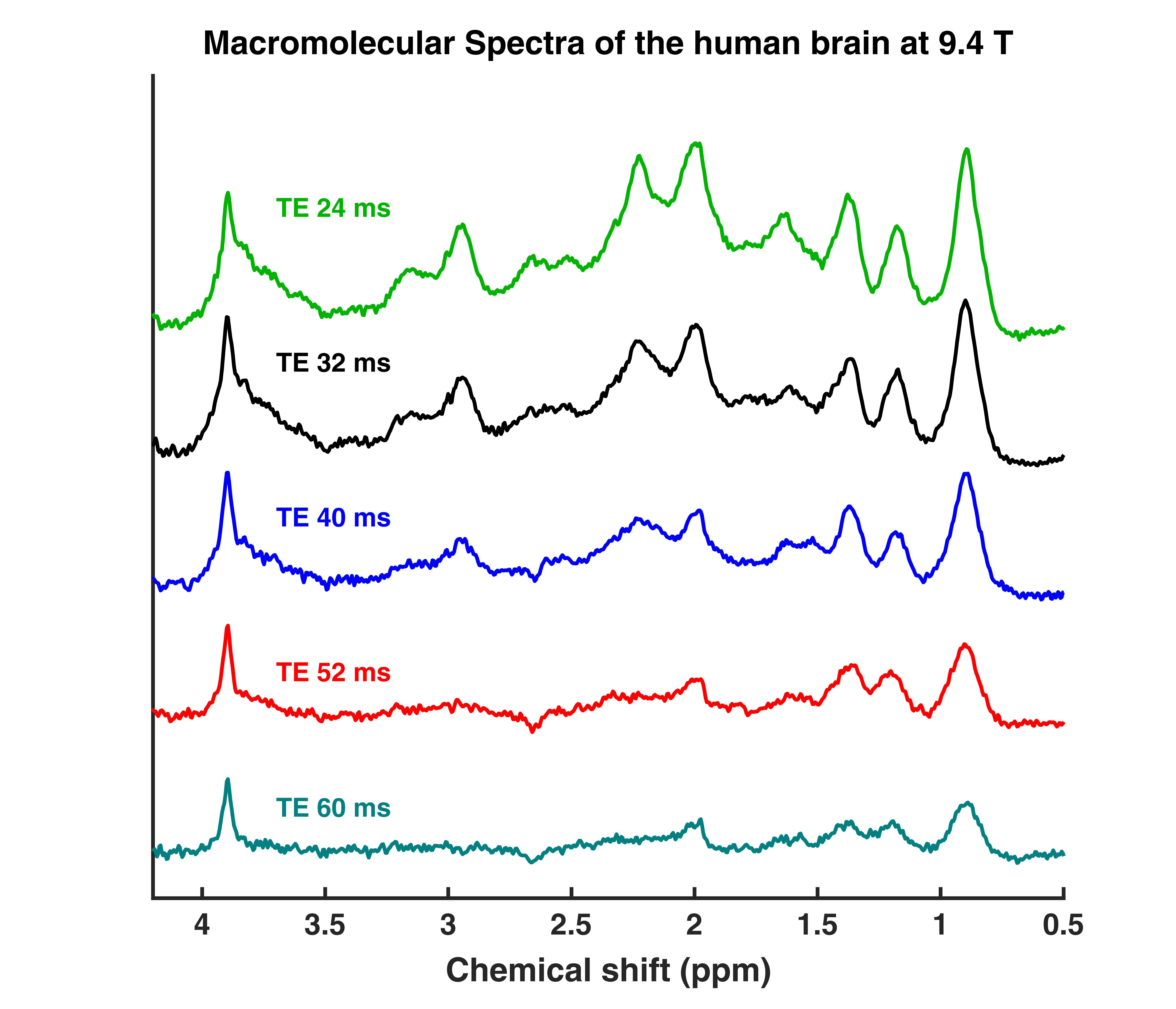
This are TE series (TE = 24,32,40,52,60 ms) DIR semi-LASER macromolecular spectra. They were measured in the occipital lobe (in GM rich and WM voxels) of the human brain. The provided data are averages from 11 subjects. Additionally, the fitsettings and sample fits are provided. An explanation on how to set up the fit settings file is provided in the Readme.MD.
| MM human brain spectra 9.4T - TE series / Fit Settings LCModel | |
|---|---|
| Developer | Tamas Borbath |
| Format | LCModel .RAW |
| Sequence | sLASER |
| License | BSD3 |
| Credit | Please cite the following publication if you use the MM human brain spectra 9.4T dataset. Murali Manohar S, Borbath T, Wright AM, Soher B, Mekle R, Henning A. T2 relaxation times of macromolecules and metabolites in the human brain at 9.4 T. Magnetic resonance in medicine. 2020;84:542–58. |
| Contact | tamas.borbath@tuebingen.mpg.de |
MM Consensus Data Collection
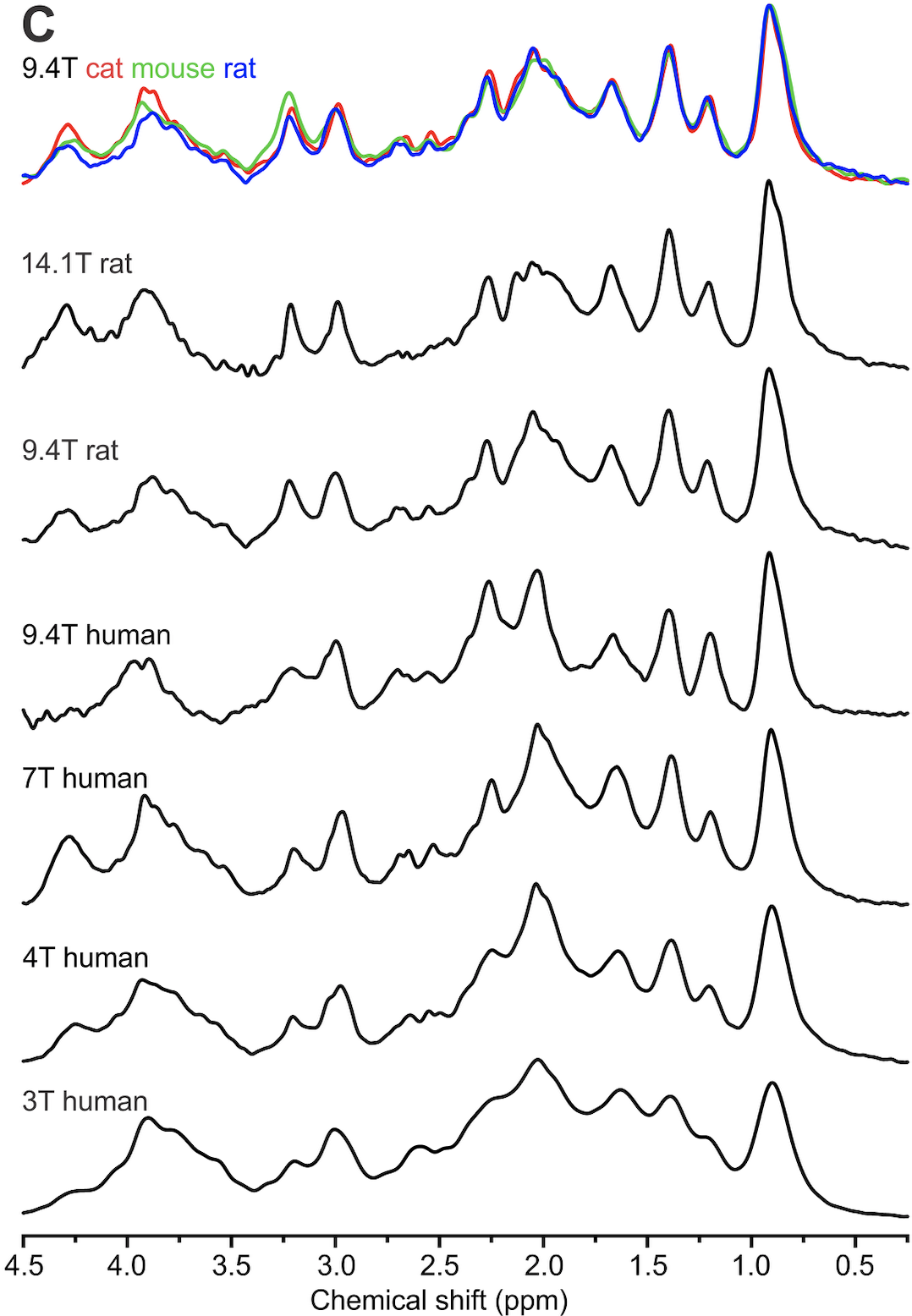
This repository contains a number of pre-processed macromolecule (metabolite-nulled) spectra in Varian format (sometimes converted from Siemens format), acquired at different field strengths from different mammals, including human. These data are represented in Figure 1 of the recently published consensus on macromolecule acquisition and handling.
| MM Consensus Data Collection | |
|---|---|
| Developer | Ivan Tkáč (4 and 7T, human; 9.4T rat, mouse, cat), Anke Henning (9.4T human), Cristina Cudalbu (14.1T, rat), Dinesh Deelchand (3T, human) |
| Format | Varian or Siemens converted in Varian |
| Sequence | STEAM, SPECIAL, sLASER |
| License | BSD3 |
| Credit | Please cite the following publications if you use data from this dataset collection: Tkáč et al., Magn Reson Med 2003; 50: 24-32 (MM 9.4T rat); Tkáč et al., Magn Reson Med 2004; 52: 478-484 (MM 9.4T mouse, cat); Tkáč et al., Magn Reson Med 2009; 62: 868-879 (MM and spectra 4T and 7T); Cudalbu et al., J Alzheimers Dis. 2012;31 Suppl 3:S101-15. doi: 10.3233/JAD-2012-120100 (MM 14.1T rat) |
| Contact | cristina.cudalbu@epfl.ch; ivan@cmrr.umn.edu |
Monte Carlo simulations of synthetic data
Monte Carlo simulation to investigate the relationship between CRLB and SD for all fitted parameters from linear combination modeling. More details about the acquisition can be found in the reference below.
| Monte Carlo simulations of synthetic data | |
|---|---|
| Developer | Karl Landheer, Christoph Juchem |
| Format | .mat file (readable in MATLAB). Synthetic spectra were generated from MARSS, fitting was performed in INSPECTOR. Both MARSS and INSPECTOR are based in MATLAB. |
| Sequence | Synthesized spectra were simulated for a TE = 20.1 ms sLASER sequence |
| License | BSD3 |
| Credit | Please cite the following publication if you use this dataset: Landheer K, Juchem C. Are Cramér-Rao Lower Bounds an Accurate Estimate for Standard Deviations in In Vivo Magnetic Resonance Spectroscopy? NMR Biomed. [In press] |
| Contact | cwj2112@columbia.edu |
MRSDB
MRSDB: A Scalable Multisite Data Library for Clinical and Machine Learning Applications of Magnetic Resonance Spectroscopy
| MRSDB | |
|---|---|
| Developer | Sam H. Jiang, BS |
| Format | CSV (from Siemens TWIX, processed in LCModel) |
| Sequence | PRESS |
| License | none |
| Credit | Center for Clinical Spectroscopy, Brigham and Women's Hospital and Harvard Medical School |
| Contact | hjiang@bwh.harvard.edu |
Considerations for event related GABA fMRS
This is the ‘Considerations for event related GABA fMRS’ dataset. It contains 3 matlab workspaces, with simulated spectra for GABA and Glutamate from a MEGA-PRESS experiment.
| Considerations for event related GABA fMRS | |
|---|---|
| Developer | Paul Mullins, PhD |
| Format | mat, MATLAB workspace |
| Sequence | MEGA-PRESS |
| License | Copyright (c) 2024, Paul Mullins. All rights reserved. Redistribution and use in source and binary forms, with or without modification, are permitted provided that the following conditions are met: 1. Redistributions of source code must retain the above copyright notice, this list of conditions and the following disclaimer. 2. Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution. 3. Neither the name of the copyright holder nor the names of its contributors may be used to endorse or promote products derived from this software without specific prior written permission. THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS ''AS IS'' AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE. |
| Credit | Citation details will be added once the manuscript is accepted |
| Contact | p.mullins@bangor.ac.uk |
Multi-site Frequency Drift MR Spectroscopy
This dataset consists of PRESS data acquired before and after running a heavy gradient duty fMRI sequence using standardized protocols for GE, Philips and Siemens scanners. 1) Pre-fMRI PRESS (TR/TE = 5000/35 ms; FA = 90°; 64 transients; no water suppression; voxel size = 2 × 2 × 2 cm3; second-order shim; scan duration = 5:20 min); 2) BOLD-weighted EPI based on the ADNI-3 protocol (Weiner et al., 2017) (TR/TE = 3000/30 ms; 197 dynamics; EPI factor/echo train length = 31/64, duration = 10 min); -3) Post-fMRI PRESS (same parameters as for pre-fMRI PRESS except 360 transients and duration = 30 min).
| Multi-site Frequency Drift MR Spectroscopy | |
|---|---|
| Developer | SCN Hui et al (Full author list bit.ly/MRSFrequencyDrift) |
| Format | SDAT, TWIX, P |
| Sequence | PRESS |
| License | This dataset is distributed freely under the Creative Commons Attribution-NonCommercial-ShareAlike license and consensus from co-authors. |
| Credit | Please cite the following publications if you use this dataset: - Hui SCN, Mikkelsen M, Zöllner HJ et al. Frequency Drift in MR Spectroscopy: An 87-scanner 3T Phantom Study, ISMRM 29th Annual Meeting, May 15-20, 2021, Vancouver - Hui SCN, Mikkelsen M, Zöllner HJ, et al. Frequency Drift in MR Spectroscopy at 3T. NeuroImage (under review) |
| Contact | stevehui@jhu.edu |
NeuroMET - SPECIAL MRS Reproducibility

Magnetic Resonance Spectroscopy Data acquired in 9 healthy volunteers using a SPECIAL Localization with three different adiabatic inversion pulses (hyperbolic secant, WURST, GOIA) at a 7T Magnetom whole-body system (Siemens Healthineers, Erlangen, Germany). Every volunteer was examined 4 times (twice on day one including a repositioning between the measurements, and twice on day two a week later without repositioning between alike measurements) to investigate the repeatability and reproducibility. Please find more details in L.T. Riemann, C.S. Aigner, S.L.R. Ellison, R. Brühl, R. Mekle, S. Schmitter, O. Speck, G. Rose, B. Ittermann, A. Fillmer, Assessment of Measurement Precision in Single Voxel Spectroscopy at 7 T: Towards Minimal Detectable Changes of Metabolite Concentrations in the Human Brain In-Vivo, Magn Reson Med DOI 10.1002/mrm.29034 (in press).
| NeuroMET - SPECIAL MRS Reproducibility | |
|---|---|
| Developer | Layla Tabea Riemann, Christoph Stefan Aigner, Stephen L.R. Ellison, Rüdiger Brühl, Ralf Mekle, Sebastian Schmitter, Oliver Speck, Georg Rose, Bernd Ittermann, Ariane Fillmer |
| Format | Siemens .dat, npy, xls |
| Sequence | SPECIAL |
| License | CC BY-NC-SA 4.0 |
| Credit | Please cite the publication DOI 10.1002/mrm.29034 as well as the data set DOI 10.5281/zenodo.5500320 if you use the NeuroMET - SPECIAL MRS Reproducibility dataset. |
| Contact | ariane.fillmer@ptb.de |
fMRS in pain
This is functional MRS (3T) during an in-scanner painful intervention. It contains 15 subjects’ baseline and functional spectra (TE = 22 ms) acquired at 3T from the human anterior cingulate cortex.
| fMRS in pain | |
|---|---|
| Developer | Jessica Archibald, Erin MacMillan & John Kramer |
| Format | Philips SDAT, data/list, nii |
| Sequence | PRESS |
| License | CC0 Creative Commons |
| Credit | Please cite the following publication if you use the fMRS in pain dataset: Archibald J., MacMillan EL., Graf C., Kozlowski P., Laule C., Kramer J.L.K. ''Metabolite activity in the anterior cingulate cortex during a painful stimulus using functional MRS''. Sci Reports 2020 |
| Contact | jessica.archibald@ubc.ca |
9.4T MR Spectra from rat hippocampus with LCModel quantification and the corresponding basis set
This repository contains the LCModel quantifications of 9.4T spectra acquired in hippocampus from 7 rats. The spectra were quntified using six different DKNTMN (spline stiffness) values (0.1, 0.25, 0.4, 0.5, 1, 5). In the folder Control_files_Basis_set you can find all the control files used in this quantification along with the corresponding basis set (metabolites/simulated using NMRScopeB from jMRUI and in vivo parameters + full MM spectrum).
| 9.4T MR Spectra from rat hippocampus with LCModel quantification and the corresponding basis set | |
|---|---|
| Developer | Dunja Simicic and Cristina Cudalbu (CIBM/EPFL) |
| Format | Varian, .RAW, .mat, .txt, .pdf, .control, .basis |
| Sequence | SPECIAL |
| License | BSD3 |
| Credit | Please cite the Zenodo resource linked below if you use the dataset (publication in progress). |
| Contact | dunja.simicic@epfl.ch; cristina.cudalbu@epfl.ch |