Macromolecules datasets
This is a list of macromolecules datasets.
7T-FID-MRSI example of metabolite-nulled macromolecules spectra
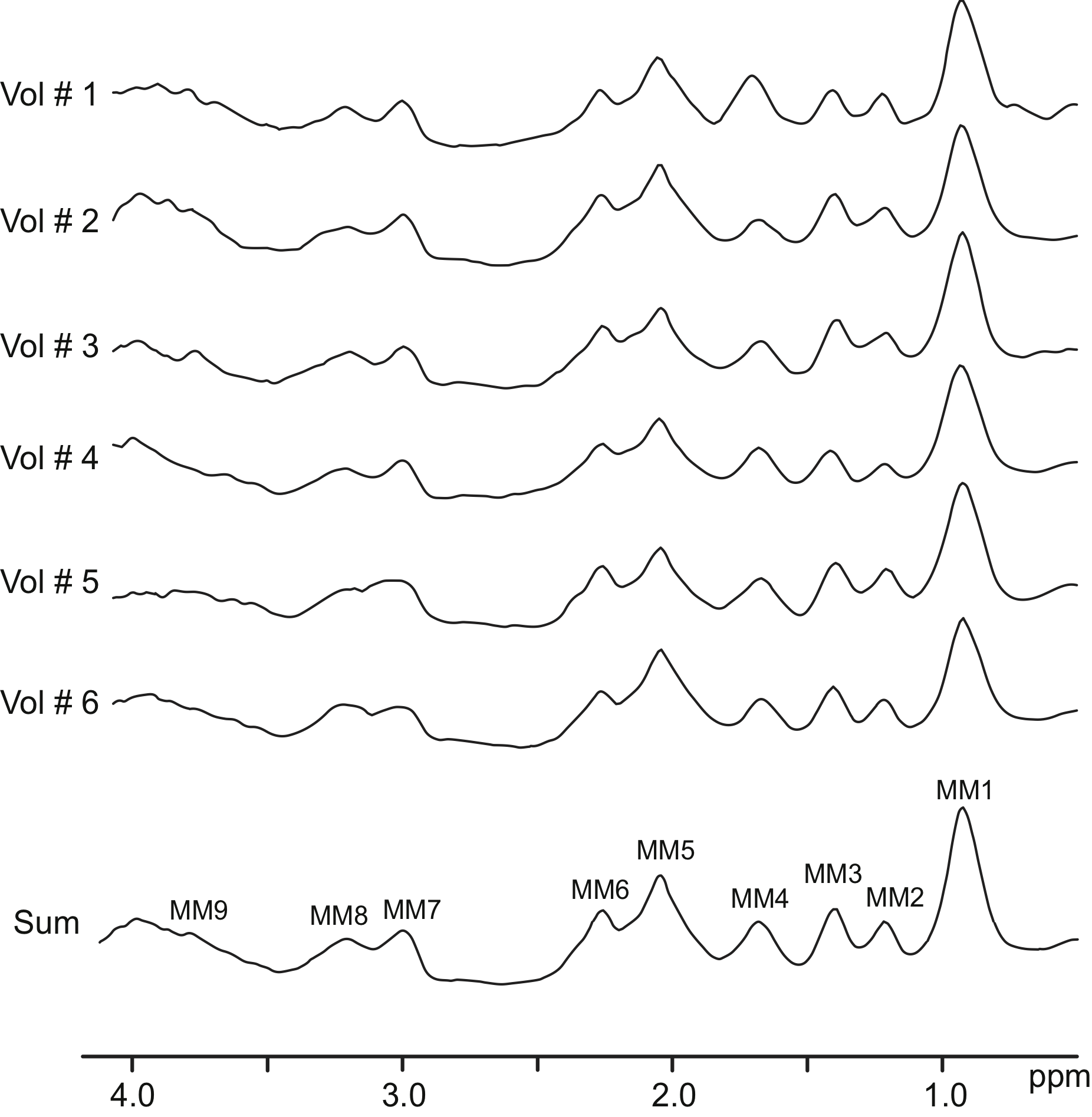
This is the example of metabolite-nulled macromolecules spectra acquired from 6 healthy young volunteers scanned at 7T using double-inversion recovery FID-MRSI
| 7T-FID-MRSI example of metabolite-nulled macromolecules spectra | |
|---|---|
| Developer | Michal Povazan, Wolfgang Bogner |
| Format | jMRUI .txt |
| Sequence | double-inversion recovery single slice FID-MRSI |
| License | BSD3 |
| Credit | Please cite Považan M, Hangel G, Strasser B, Gruber S, Chmelik M, Trattnig S, et al. Mapping of brain macromolecules and their use for spectral processing of 1H-MRSI data with an ultra-short acquisition delay at 7T. Neuroimage. 2015;121:126–35if you use this dataset. |
| Contact | mpovaza1@jhmi.edu; wolfgang.bogner@meduniwien.ac.at |
Collection of brain macromolecules spectra measured at 3 T
Metabolite-nulled macromolecules spectra acquired from 10 healthy young volunteers scanned at 3 T using double-inversion recovery semi-LASER sequence with 8 echo times (20 - 62 ms) from two brain regions (prefrontal and occipital). Voxel-specific segmented brain values are included in Excel spreadsheet. More details about the acquisition can be found in the reference below.
| Collection of brain macromolecules spectra measured at 3 T | |
|---|---|
| Developer | Karl Landheer, Martin Gajdošík, Michael Treacy, Christoph Juchem |
| Format | INSPECTOR .mat, jMRUI .txt |
| Sequence | double-inversion recovery sLASER |
| License | BSD3 |
| Credit | Please cite this publication if you use this dataset: Landheer K, Gajdošík M, Treacy M, Juchem C. Concentration and effective T 2 relaxation times of macromolecules at 3T. Magn Reson Med. 2020 Nov;84(5):2327-2337 |
| Contact | cwj2112@columbia.edu |
MM human brain spectra 9.4T - TE series / Fit Settings LCModel
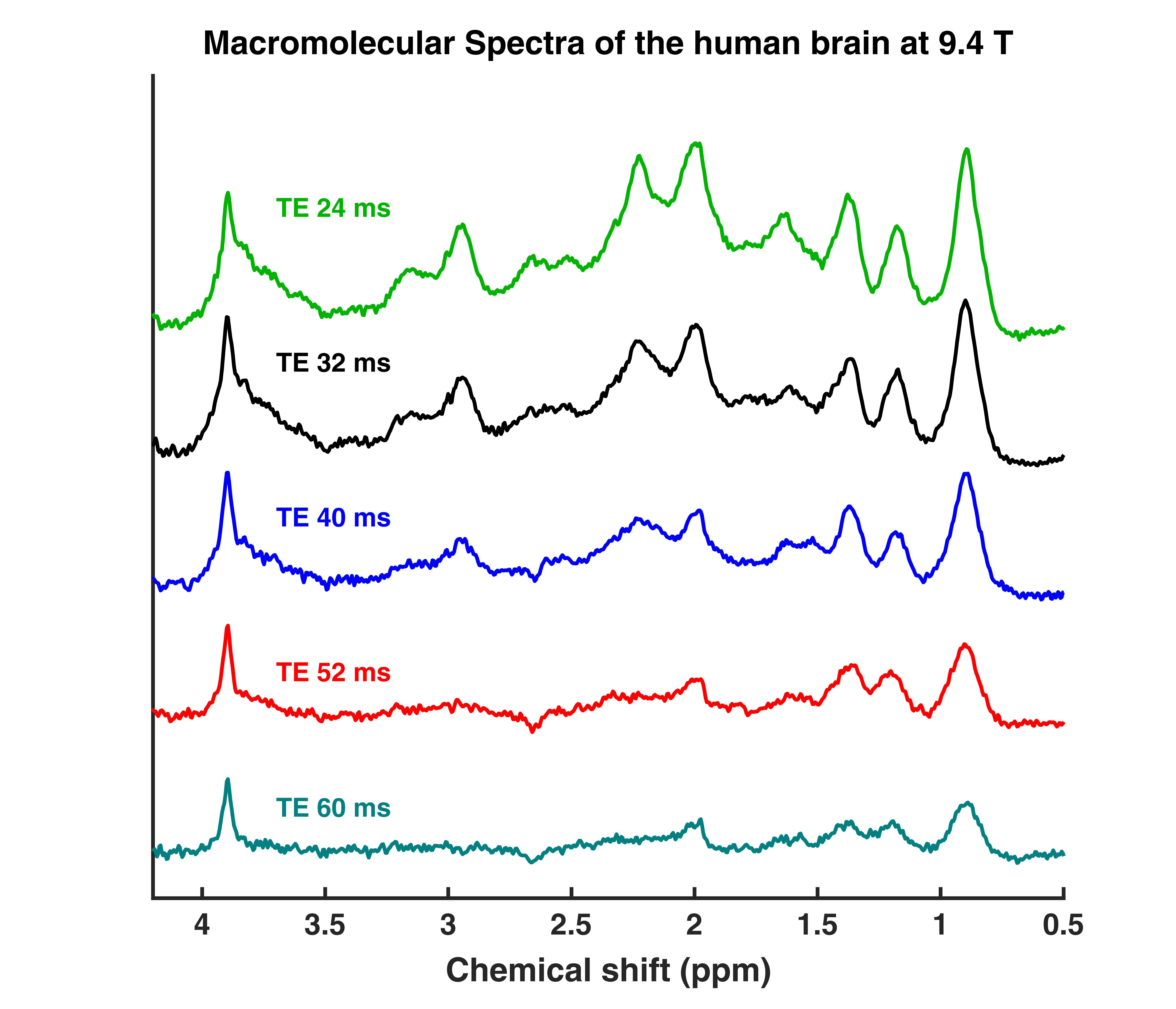
This are TE series (TE = 24,32,40,52,60 ms) DIR semi-LASER macromolecular spectra. They were measured in the occipital lobe (in GM rich and WM voxels) of the human brain. The provided data are averages from 11 subjects. Additionally, the fitsettings and sample fits are provided. An explanation on how to set up the fit settings file is provided in the Readme.MD.
| MM human brain spectra 9.4T - TE series / Fit Settings LCModel | |
|---|---|
| Developer | Tamas Borbath |
| Format | LCModel .RAW |
| Sequence | sLASER |
| License | BSD3 |
| Credit | Please cite the following publication if you use the MM human brain spectra 9.4T dataset. Murali Manohar S, Borbath T, Wright AM, Soher B, Mekle R, Henning A. T2 relaxation times of macromolecules and metabolites in the human brain at 9.4 T. Magnetic resonance in medicine. 2020;84:542–58. |
| Contact | tamas.borbath@tuebingen.mpg.de |
MM Consensus Data Collection
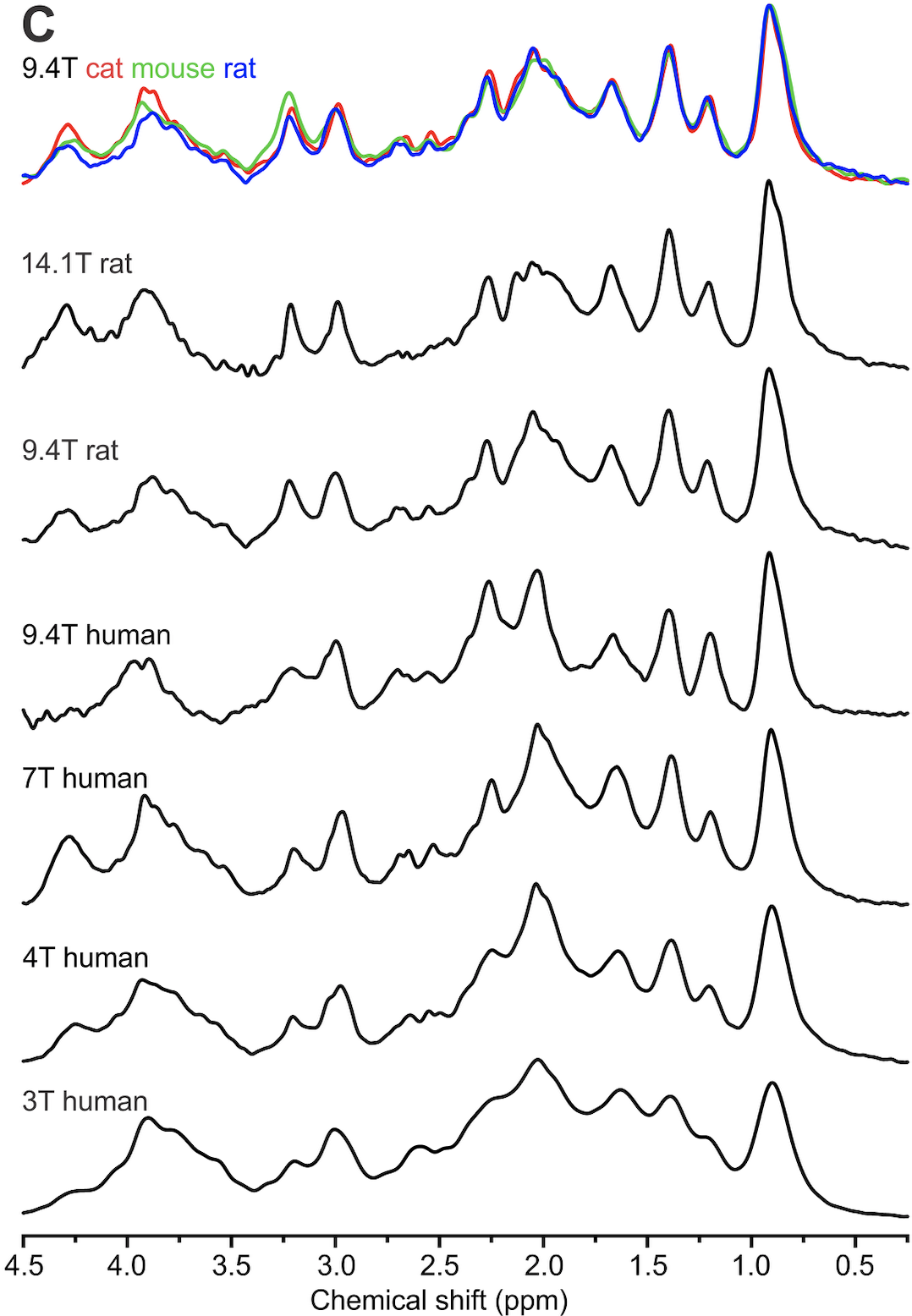
This repository contains a number of pre-processed macromolecule (metabolite-nulled) spectra in Varian format (sometimes converted from Siemens format), acquired at different field strengths from different mammals, including human. These data are represented in Figure 1 of the recently published consensus on macromolecule acquisition and handling.
| MM Consensus Data Collection | |
|---|---|
| Developer | Ivan Tkáč (4 and 7T, human; 9.4T rat, mouse, cat), Anke Henning (9.4T human), Cristina Cudalbu (14.1T, rat), Dinesh Deelchand (3T, human) |
| Format | Varian or Siemens converted in Varian |
| Sequence | STEAM, SPECIAL, sLASER |
| License | BSD3 |
| Credit | Please cite the following publications if you use data from this dataset collection: Tkáč et al., Magn Reson Med 2003; 50: 24-32 (MM 9.4T rat); Tkáč et al., Magn Reson Med 2004; 52: 478-484 (MM 9.4T mouse, cat); Tkáč et al., Magn Reson Med 2009; 62: 868-879 (MM and spectra 4T and 7T); Cudalbu et al., J Alzheimers Dis. 2012;31 Suppl 3:S101-15. doi: 10.3233/JAD-2012-120100 (MM 14.1T rat) |
| Contact | cristina.cudalbu@epfl.ch; ivan@cmrr.umn.edu |