Basis sets
This is a list of basis sets.
Basis sets for PRESS, STEAM and sLASER for multiple vendors
These basis sets were generated using the exact waveforms, timings and 128^3 spatial points to appropriately accommodate the sidebands for each vendor/sequence combination. More details about the acquisition can be found in the reference below.
| Basis sets for PRESS, STEAM and sLASER for multiple vendors | |
|---|---|
| Developer | Karl Landheer, Kelley Swanberg, Christoph Juchem |
| Format | Both .RAW and .mat (INSPECTOR, readable in MATLAB) are provided. All files were generated using MATLAB |
| Sequence | PRESS, sLASER, STEAM |
| License | BSD3 |
| Credit | Please cite the following publication if you use this dataset: Landheer K, Swanberg, K, Juchem C. Magnetic resonance Spectrum simulator (MARSS), a novel software package for fast and computationally efficient basis set simulation. NMR Biomed. 2019, e4219. doi.org/10.1002/nbm.4129 |
| Contact | cwj2112@columbia.edu |
FID-MRSI 3T and 7T basis sets with measured MM
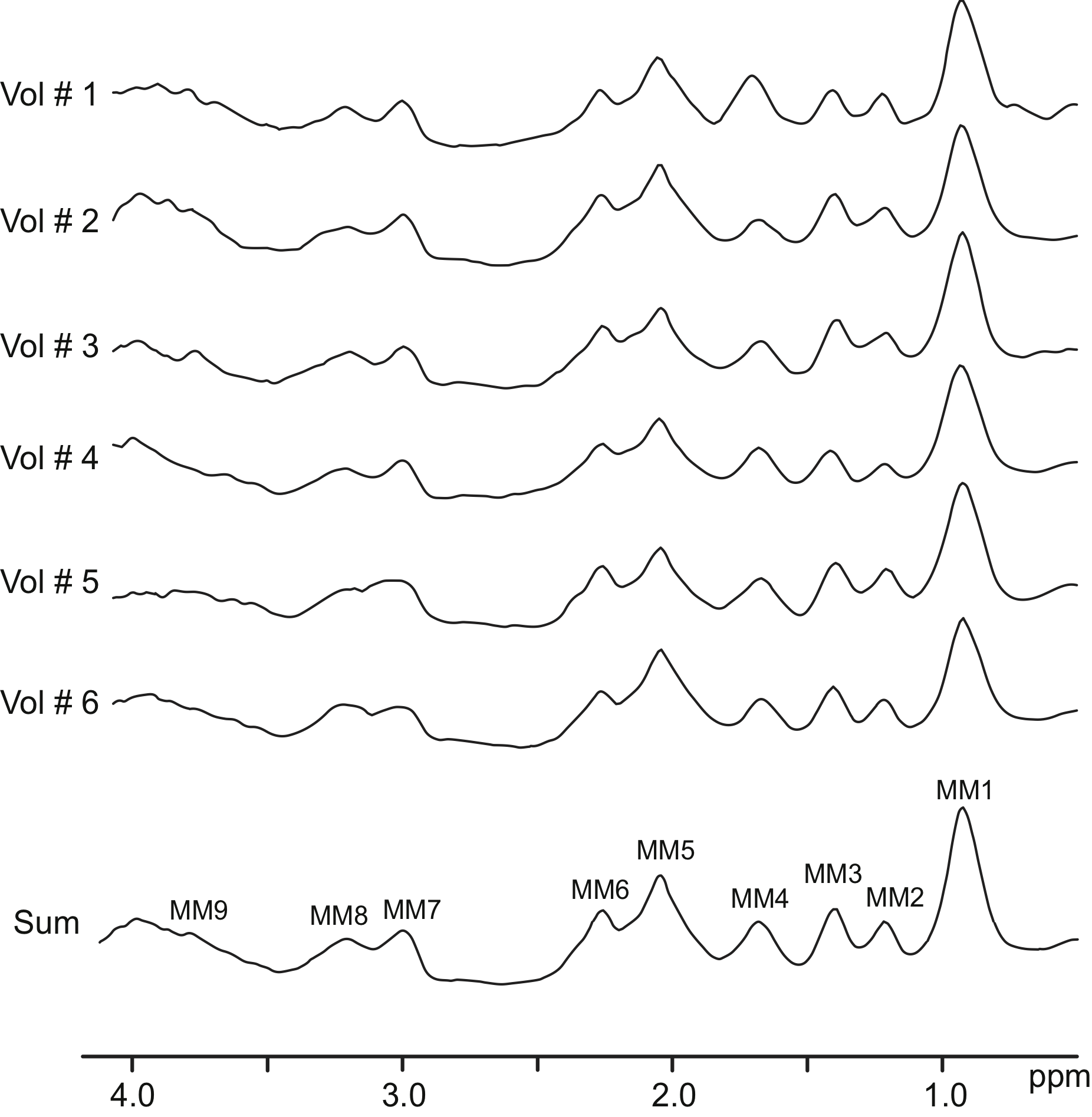
These are FID-MRSI basis sets with metabolite information simulated in NMR-Scope B and measured MM (see Publication Povazan et al, NI, 2015)
| FID-MRSI 3T and 7T basis sets with measured MM | |
|---|---|
| Developer | Wolfgang Bogner, Gilbert Hangel, Michal Povazan, Bernhard Strasser |
| Format | LCModel .basis and .RAW |
| Sequence | FID-MRSI with acquisition delay of 0 ms / 1.3 ms / 1.5 ms |
| License | BSD3 |
| Credit | Please consult the authors how to cite these basis sets |
| Contact | mpovaza1@jhmi.edu; wolfgang.bogner@meduniwien.ac.at |
Neonatal 3T PRESS Basis Sets (MARBLE)
High resolution basis sets for neonatal MRS with vendor-provided SV PRESS at TE=60ms and TE=288ms, simulated using Vespa and used in the MARBLE study (see Lally P.J., Montaldo P. et al. Lancet Neuro 2019).
| Neonatal 3T PRESS Basis Sets (MARBLE) | |
|---|---|
| Developer | Peter J Lally, PhD |
| Format | .basis .RAW .in .pdf |
| Sequence | PRESS (TE=60ms, TE=288ms) |
| License | BSD3 |
| Credit | Please cite the publication mentioned below if you use these basis sets. |
| Contact | p.lally [at] imperial.ac.uk |